
Food Quality and Safety
The main objective of the Food & Quality Research Group is the development of analytical approaches based on the combination of molecular biology (mainly DNA based methodology) and nano and microfabrication technology in order to provide the food industry and control laboratories with reliable analytical tools.
Following this objective, the methodology is based on working on very specific analytical needs and on using a modular approach for each of the steps of the analytical process. This approach, help us to evaluate and to choose the best method in each case, to have a sound integrated final product, and at the same time a wide-range of intermediate products that can be used by themselves to solve specific analytical challenges.
Figure 1 summarizes the overall approach and research lines. Our topics of interest involve the detection of foodborne pathogens, the detection of allergenic ingredients in food products and food authenticity.
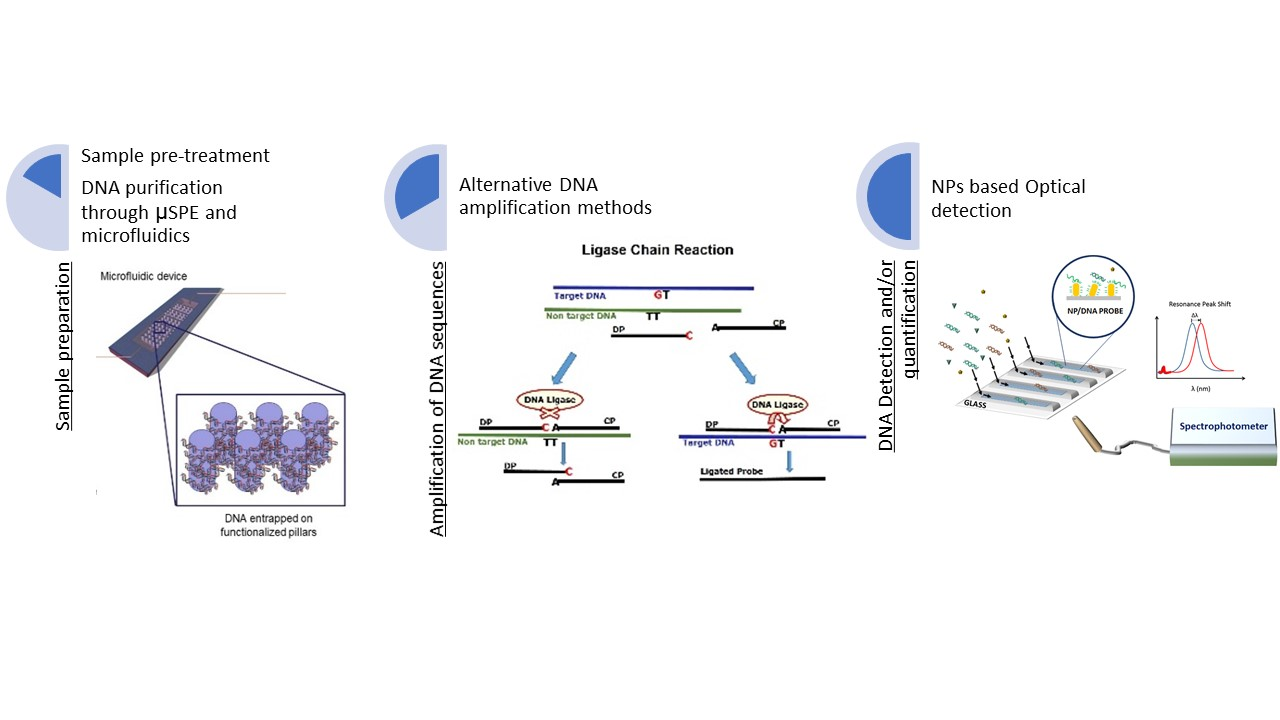
Research Lines:
- Sample preparation:
Sample preparation is the series of steps required to transform a sample to a form suitable for analysis, the reliability of the conclusions drawn from food analysis greatly depends upon on this step. We work on: (i) the development of pre-treatment steps in order to overcome some of the limitations associated with food analysis and (ii) on the development of tailored, miniaturized, automatized and faster sample preparation techniques. Microscale solid phase extraction (µSPE) is used for on-chip DNA extraction and purification, being possible to put in contact a higher volume of initial binding material with the solid phase and recover the DNA in a lower volume during the elution phase. This feature allows to concentrate the DNA when minute amounts are present in the sample (e.g. olive oil, wine), for complex matrixes such as processed foodstuff and for environmental samples (e.g. water samples).
- Alternative DNA amplification methods:
Food & Quality Safety Research Group is working on new amplification techniques, in their combination with NPs and on the evaluation of DNA based analytical methods for food analysis. We work on isothermal amplification techniques, such as Loop-Mediated Isothermal Amplification (LAMP), and Recombinase Polymerase Amplification (RPA), specially interesting for miniaturization purposes. Other alternative techniques currently being used include Ligation Chain Reaction (LCR) which allows to distinguish very closely related organisms and high similar DNA sequences.
- Nanoparticle-assisted DNA analysis:
The use of nanomaterials for DNA analysis has the potential of providing increased sensitivity, multiplexing capabilities, and reduced costs. Exploiting the features of nanoparticles (NPs) is considered to be a good alternative to foster the potential of diagnostics and analytical method development. NPs, such as gold NPs (AuNPs) and gold nanorods (AuNRs) are being used for DNA detection taking advantage of their optical properties.
Projects
Publications
-
Evaluation of the Antimicrobial Activity of Chitosan Nanoparticles against Listeria monocytogenes
POLYMERS, 2023Green-Based Modifications of Polyethylene Surfaces Enhancing Antifouling Properties in Water-Related Systems
ADVANCED MATERIALS TECHNOLOGIES, 2023Integrated Approach from Sample-to-Answer for Grapevine Varietal Identification on a Portable Graphene Sensor Chip
ACS SENSORS, 2023 -
A novel portable label-free electrochemical immunosensor for ultrasensitive detection of Aeromonas salmonicida in aquaculture seawater
ANALYTICAL AND BIOANALYTICAL CHEMISTRY, 2022Programmable graphene-based microfluidic sensor for DNA detection
SENSORS AND ACTUATORS B-CHEMICAL, 2022Rapid Same-Day Detection of Listeria monocytogenes, Salmonella spp., and Escherichia coli O157 by Colorimetric LAMP in Dairy Products
FOOD ANALYTICAL METHODS, 2022Towards on-site detection of gluten-containing cereals with a portable and miniaturized prototype combining isothermal DNA amplification and naked eye detection
MICROCHEMICAL JOURNAL, 2022Development of a real-time PCR assay with an internal amplification control for the detection of spoilage fungi in fruit preparations
FOOD CONTROL, 2022Development of a Panfungal Recombinase Polymerase Amplification (RPA) Method Coupled with Lateral Flow Strips for the Detection of Spoilage Fungi.
FOOD ANALYTICAL METHODS, 2022Naked-eye detection strategies coupled with isothermal nucleic acid amplification techniques for the detection of human pathogens
COMPREHENSIVE REVIEWS IN FOOD SCIENCE AND FOOD SAFETY, 2022Sensing Alzheimer's Disease Utilizing Au Electrode by Controlling Nanorestructuring
CHEMOSENSORS, 2022Rapid Same-Day Detection of Listeria monocytogenes, Salmonella spp., and Escherichia coli O157 by Colorimetric LAMP in Dairy Products.
FOOD ANALYTICAL METHODS 15, pages2959–2971, 2022Versatile and Low-Cost Fabrication of Modular Lock-and-Key Microfluidics for Integrated Connector Mixer Using a Stereolithography 3D Printing
MICROMACHINES, 2022The Protein-Templated Synthesis of Enzyme-Generated Aptamers
ANGEWANDTE CHEMIE-INTERNATIONAL EDITION, 2022Combination of Recombinase Polymerase Amplification with SYBR Green I for naked-eye, same-day detection of Escherichia coli O157:H7 in ground meat
FOOD CONTROL, 2022Dual-Mode Gold Nanoparticle-Based Method for Early Detection of Acanthamoeba
INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES, 2022 -
A multivalent aptamer-based electrochemical biosensor for biomarker detection in urinary tract infection
ELECTROCHIMICA ACTA, 2021Amplified plasmonic and microfluidic setup for DNA monitoring
MICROCHIMICA ACTA, 2021Suitability of the MinION long read sequencer for semi-targeted detection of foodborne pathogens
Analytica Chimica Acta Volume 1184, 1 November 2021, 339051, 2021Single-use microfluidic device for purification and concentration of environmental DNA from river water
TALANTA, 2021Application of Omics-based Miniaturized Systems in Food Quality and Safety
Book: Foodomics: Omic Strategies and Applications in Food Science (2021), Chapter 9, pp. 222 – 256 ISBN: 978-1-78801-884-5, 2021Evaluation of simple sequence repeats (SSR) and single nucleotide polymorphism (SNP)-based methods in olive varieties from the Northwest of Spain and potential for miniaturization
FOOD CHEMISTRY: MOLECULAR SCIENCES, 2021Faster monitoring of the invasive alien species (IAS) Dreissena polymorpha in river basins through isothermal amplification
SCIENTIFIC REPORTS, 2021Gold nanoparticle-assisted plasmonic enhancement for DNA detection on a graphene-based portable surface plasmon resonance sensor
NANOTECHNOLOGY, 2021Influence of the Electrolyte Salt Concentration on DNA Detection with Graphene Transistors
BIOSENSORS-BASEL, 2021Terminal deoxynucleotidyl transferase-mediated formation of protein binding polynucleotides
NUCLEIC ACIDS RESEARCH, 2021Gold Nanoframe Array Electrode for Straightforward Detection of Hydrogen Peroxide
CHEMOSENSORS, 2021Dual colorimetric strategy for specific DNA detection by nicking endonuclease-assisted gold nanoparticle signal amplification
ANALYTICAL AND BIOANALYTICAL CHEMISTRY, 2021 -
Evaluation and implementation of commercial antibodies for improved nanoparticle-based immunomagnetic separation and real-time PCR for faster detection of Listeria monocytogenes
JOURNAL OF FOOD SCIENCE AND TECHNOLOGY-MYSORE, 2020A smart microfluidic platform for rapid multiplexed detection of foodborne pathogens
FOOD CONTROL, 2020ZnO-Nanorod processed PC-SET as the light-harvesting model for plasmontronic fluorescence Sensor
SENSORS AND ACTUATORS B-CHEMICAL, 2020Chapter 2. New techniques in environment monitoring.
Climate Change and Marine and Freshwater Toxins. De Gruyter, 2020Photoelectrochemical Detection of beta-amyloid Peptides by a TiO2 Nanobrush Biosensor
IEEE SENSORS JOURNAL, 2020Optimized sample treatment, combined with real-time PCR, for same-day detection of E. coli O157 in ground beef and leafy greens
Food Control 108 (2020): 106790., 2020Photoelectrochemical Detection of β-amyloid Peptides by a TiO2 Nanobrush Biosensor
IEEE Sensors Journal 2020. PP(99):1-1, 2020Facile Bacterial Cellulose Nanofibrillation for the Development of a Plasmonic Paper Sensor
ACS BIOMATERIALS SCIENCE & ENGINEERING, 2020Multifuntional Gold Nanoparticles for the SERS Detection of Pathogens Combined with a LAMP-in-Microdroplets Approach
MATERIALS, 2020 -
Gold Nanostars for the Detection of Foodborne Pathogens via Surface-Enhanced Raman Scattering Combined with Microfluidics
ACS APPLIED NANO MATERIALS, 2019Specific detection of viable Salmonella Enteritidis by phage amplification combined with qPCR (PAA-qPCR) in spiked chicken meat samples
FOOD CONTROL, 2019Combination of Immunomagnetic Separation and Real‐Time Recombinase Polymerase Amplification (IMS‐qRPA) for Specific Detection of Listeria monocytogenes in Smoked Salmon Samples
Journal of food science 84 (7) (2019): 1881-1887, 2019Amplification-free SERS analysis of DNA mutation in cancer cells with single-base sensitivity
NANOSCALE, 2019Attomolar Label-Free Detection of DNA Hybridization with Electrolyte-Gated Graphene Field-Effect Transistors
ACS SENSORS, 2019
